A reference database for splicing variations across normal tissues and cancer in human
Explore splicing variations in:
What is MAJIQlopedia?
MAJIQlopedia is a database with a built-in visual summary interface for all splicing variations found in a large set of normal and cancer human samples. These local splicing variations (LSVs), all quantied by the MAJIQ algorithm (hyperlink to MAJIQ v2 paper) include annotated and unannotated splice junctions, complex LSVs (involving more than two splice junctions) and intron retention. A more detailed denition of LSVs and visual examples of those can be found in the FAQs.
How to use:
Choose whether you want to visualize splicing variations in normal tissues or in cancer, then enter the name of the gene (Gene symbol or EnsemblID) you want to query. The search will return all genes matching your query for which splicing variations were quantified. Selecting the gene of interest will take you to a page showing a splicegraph with all alternative splice junctions found in the gene at the top of the page and a visual representation of the distributions of how much each junction in an local splicing variation (LSV) is being used in different tissues. The junction usage value is given in “percent spliced-in” (PSI) and ranges between 0 and 1.
At the top of the result page, you can choose the reference transcript to display. The reference transcript is aligned to the splicegraph built by MAJIQ from observed sequencing reads.
Splicegraphs can be toggled in and out of view using the “Toggle Splice Graph” option a the very top of the page. Tissues that appear in the junction usage quantification plots can be selected individually using the “Plot Options” option at the very top of the page.
Detailed methods and case usage examples are provided in a manuscript currently under review.
What are local splicing variations (LSVs)?
Each local splicing variation (LSV) consists of two or more splice junctions stemming from a given reference exon. A target LSV consists of two or more junctions connecting to a reference exon downstream while a single source LSV involves multiple junctions stemming out of an upstream exon as illustrated below
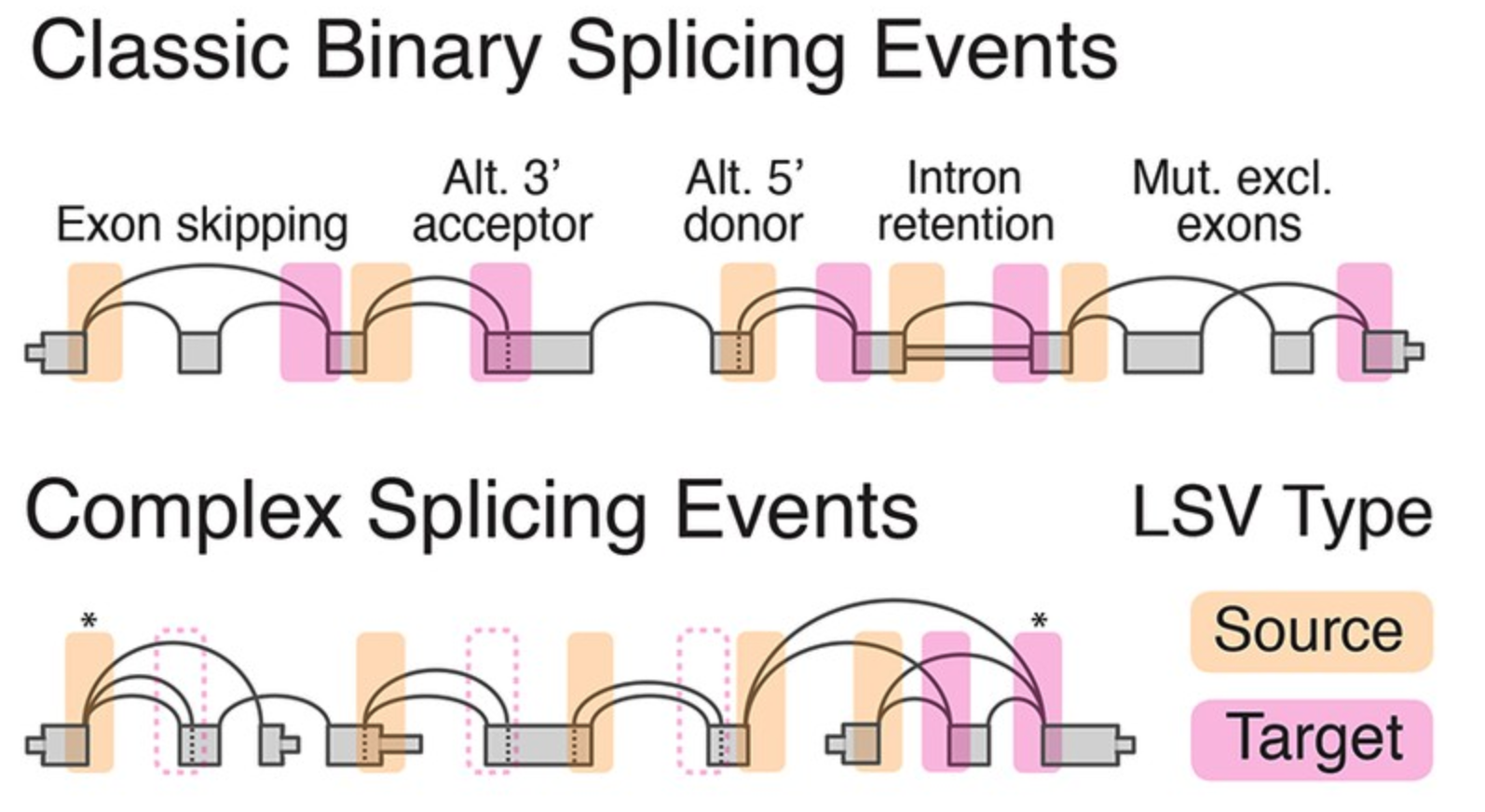
(See more details HERE)
What data does MAJIQlopedia include?
The MAJIQlopedia was built using short-read RNA-Seq data combined from several large scale efforts and smaller studies including:
- GTEX (GTEx Consortium, 2013, Nat Genet)
- Blueprint (Adams et al., 2012, Nat Biotech)
- Leucegene (MacRae et al., 2013, PLoS One)
- TCGA (The Cancer Genome Atlas Research Network, 2013, Nature Genet)
- TARGET (Ma et al., 2018, Nature)
- Beat-AML (Tyner et al., 2018, Nature)
- CBTN (Lilly et al., 2023, Neoplasia)
- T cells (Ranzani et al., 2015, Nat Immunol)
- bone cells (Moriarity et al., 2015, Nat Genet [https://www.nature.com/articles/ng.3293]; Roforth et al., 2014, Bone)
Why/when should I use MAJIQlopedia?
There are many databases that list splice junctions across a vast array of experiments. The advantage of MAJIQlopedia lies in the accurate quantification and matching visualization of those junctions, combined with intron retention information which is typically missed in those databases. Rather than getting only junction read counts, MAJIQlopedia also translates those into splice graphs, LSVs, and matching PSI values. Some use cases include:
- User is interested to know if a splice variant found in patient or specific experimental condition can be found in cancer cell lines for further analysis/manipulation.
- User is interested to know if a splice variant is unique to a specific condition (e.g. cancer type) or in which tissues such a splice variant is being used.
- User is interested in how a splice junction "fits" within a gene splice graph and the splice variations around it.
- User wants to know what splicing variations exist in their gene(s) of interest.
Can I select the threshold for the number of reads required for a splicing variation to appear in the output of MAJIQlopedia?
At the moment it is not possible to change coverage stringency for the local splicing variations (LSVs) that appear in MAJIQlopedia. The database was built requiring a minimum of 10 reads covering each LSV in order for the event to appear. This stringent threshold was selected in order to filter out low-confidence events. Depending on demand, we might consider adding the option to further increase the required number of reads for an LSV to appear.
Why do some splicing events/junctions appear twice in the database?
Because MAJIQ quantifies alternative splicing locally, the splice junctions composing an alternative splicing event will appear from both "source" (quantified from the exon in 5′) and "target" (quantified from the exon in 3′) local splicing variations. Please see the documentation for a complete description of how MAJIQ quantifies alternative splicing.
Why are some columns in the quantification plots empty?
Empty columns indicate that the junction could not be quantified in the tissue. This often occurs when the gene is expressed at low levels in a tissue, when the isoforms used by a tissue do not involved the reference exon that the local splicing variation originates from, or if the depth of the sequencing for a tissue was too low to allow for quantification by MAJIQ.
Can MAJIQlopedia be used to perform differential splicing analysis?
The data provided in MAJIQlopedia was not designed to be used in differential splicing analyses. The list of splicing changes in a cancer of interest relative to its tissue of origin can be achieved by running the publicly available MAJIQ Heterogen module (Vaquero-Garcia et al., Nature Commun, 2023) on datasets of interest for this specific purpose.
Why do exon numbers differ between the annotated transcripts and the MAJIQ splicegraph?
Exon numbers on annotated transcripts correspond to exon labeling on the selected annotated transcript. On the MAJIQ splicegraph, exons are numbered from the 5′ of the transcript to the 3′ end. It is possible that MAJIQ will detect exons that do not appear in the selected reference transcript, which will shift the labeling between the two representations.
Download Summary Data files
Here we provide the raw data used to generate the junction quantification plots provided in the webtool. psi.tsv files contain the median PSI values for each variable junction in all of 86 normal tissues or 41 cancers; stdev.tsv files contain the standard deviation for each of the PSI values provided in the psi.tsv files.
Please note that the absolute number of splice junctions reported for each tissue/cancer type depends on the number of samples and sequencing depth of each dataset. These parameters vary between tissue and cancer types. Comparing the total number of variable junctions found in different tissues/cancers is therefore discouraged.
MAJIQlopedia: an encyclopedia of RNA splicing variations in human tissues and cancer
Mathieu Quesnel-Vallières, San Jewell, Kristen W Lynch, Andrei Thomas-Tikhonenko, Yoseph Barash
Nucleic Acids Research, Volume 52, Issue D1, 5 January 2024, Pages D213–D221, https://doi.org/10.1093/nar/gkad1043
Published: 11 November 2023
View Article»Questions?
For general help with this tool, or to contact the authors, please use the form below
Alternatively, for potentially faster replies, we recommend using the MAJIQ board (https://groups.google.com/g/majiq_voila/), please remember to tag your questions with the "MAJIQlopedia" label
